Dorado v1.0.0 and the v5.2.0 basecalling models
Last week, ONT’s London Calling conference brought the release of Dorado v1.0.0. While I think of Dorado as a basecaller, ONT keeps adding other features: it aligns, trims, corrects, demultiplexes, polishes and (as of this release) calls variants. But for this post, I’ll focus only on basecalling.
A new version of Dorado is nice, but what really caught my attention was that it came with new DNA basecalling models: version 5.2.0. It’s been a full year since the previous models (v5.0.0), so I’m hoping for a boost in read and assembly accuracy. That’s what I’ll test in this blog post!
Model architectures
For both v5.0.0 and v5.2.0, the models follow the same basic architecture: hac uses LSTMs, and sup uses transformers.
The hac model has grown from five alternating-direction LSTM layers in v5.0.0 to seven in v5.2.0, making it 37% larger by parameter count. Dorado has seen many performance improvements in recent versions, and I suspect those optimisations gave ONT the headroom to increase model size while keeping hac basecalling quick enough on existing hardware.
The sup model architecture appears unchanged from v5.0.0 to v5.2.0, so differences there will come from training data and weights.
I didn’t test the fast models here, but their architecture also appears unchanged from v5.0.0 to v5.2.0.
Methods
I used the same dataset as in the Autocycler preprint. Briefly: five bacterial isolates with high-quality reference genomes (Illumina-polished), sequenced (along with other isolates) on a PromethION flowcell (~132 Gbp total run yield).
I basecalled the full run four times:1
hac@5.0.0: Dorado v0.9.5 withdna_r10.4.1_e8.2_400bps_hac@v5.0.0hac@5.2.0: Dorado v1.0.0 withdna_r10.4.1_e8.2_400bps_hac@v5.2.0sup@5.0.0: Dorado v0.9.5 withdna_r10.4.1_e8.2_400bps_sup@v5.0.0sup@5.2.0: Dorado v1.0.0 withdna_r10.4.1_e8.2_400bps_sup@v5.2.0
To assess read accuracy, I simply aligned reads to their reference genome and calculated the identity of the alignments.2
To assess assembly accuracy, I made six non-overlapping 50× read subsets per isolate (30 total). Each was assembled with Autocycler (with some manual curation to ensure that all small plasmids were included), and errors were counted using the assessment script from the Autocycler preprint.
Speed performance
Basecalling was done on an H100 GPU on the University of Melbourne’s Spartan cluster.
| Dorado version | Model | Model parameters |
Time (h:m) | Speed (samples/sec) |
|---|---|---|---|---|
| 0.9.5 | hac@5.0.0 |
6,425,328 | 2:39 | 1.91×108 |
| 1.0.0 | hac@5.2.0 |
8,790,768 | 3:18 | 1.53×108 |
| 0.9.5 | sup@5.0.0 |
78,718,162 | 25:06 | 2.01×107 |
| 1.0.0 | sup@5.2.0 |
78,718,162 | 18:45 | 2.69×107 |
Hac basecalling got ~20% slower with the new model, likely due to its increased size.
Sup basecalling got ~33% faster, which surprised me since the model size is unchanged. This may be due to Dorado optimisations, though I didn’t spot anything obvious in the release notes between v0.9.5 and v1.0.0. Or it could be due to cluster factors like shared node usage. These benchmarks weren’t tightly controlled, so take the timing results with a grain of salt.
Read accuracy
The violin plots below show the read identity distributions (higher is better).3 The line inside each violin indicates the median.
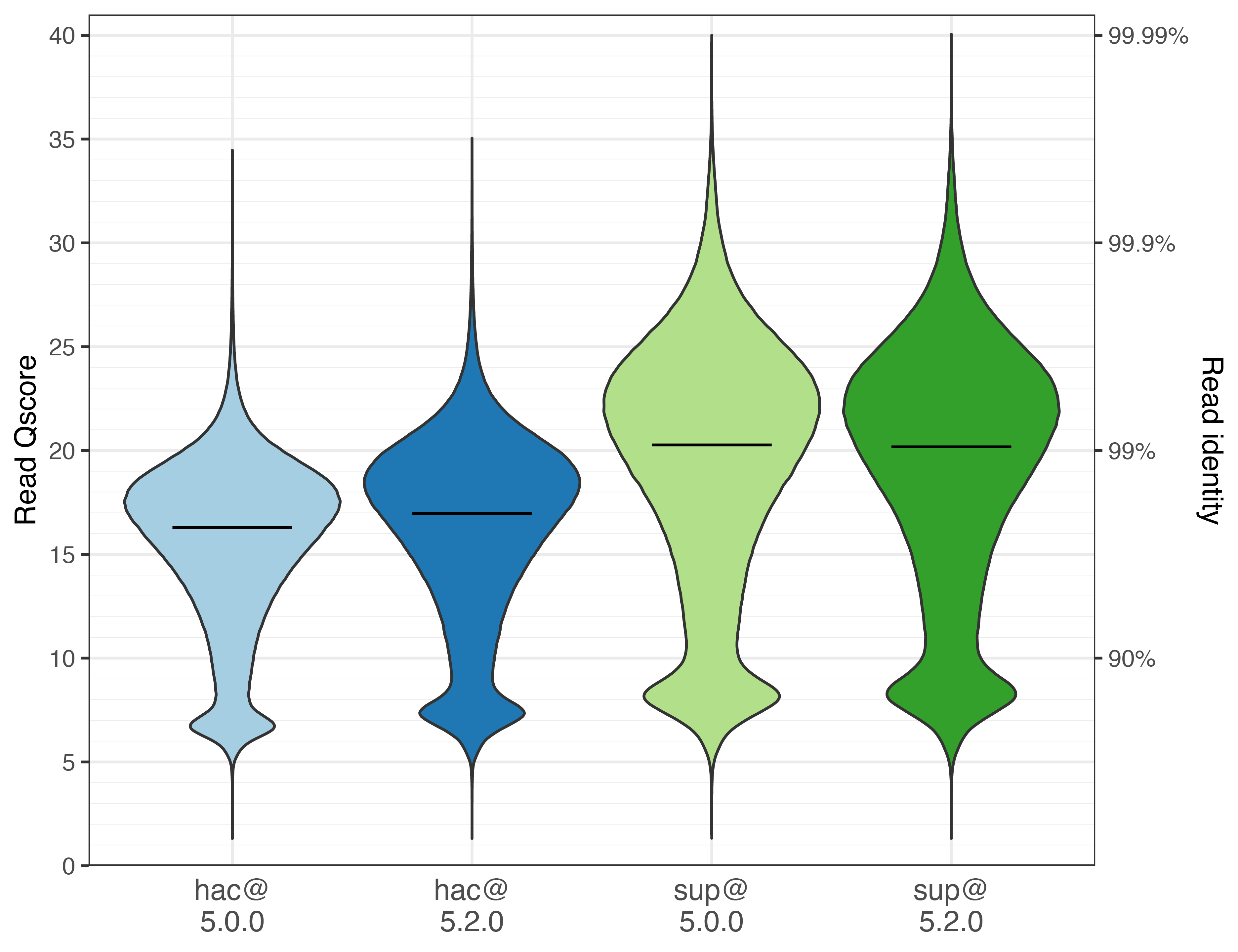
For hac, median identity increased from Q16.3 (97.65%) with hac@5.0.0 to Q17.0 (97.99%) with hac@5.2.0, which corresponds to ~15% fewer read errors.
For sup, there was little change. The newer sup@5.2.0 reads actually had a slightly lower median than the older sup@5.0.0 reads, but the difference was small.
Assembly accuracy
The boxplots below show the number of assembly errors (lower is better). The line in each box shows the median, and whiskers span the full range.
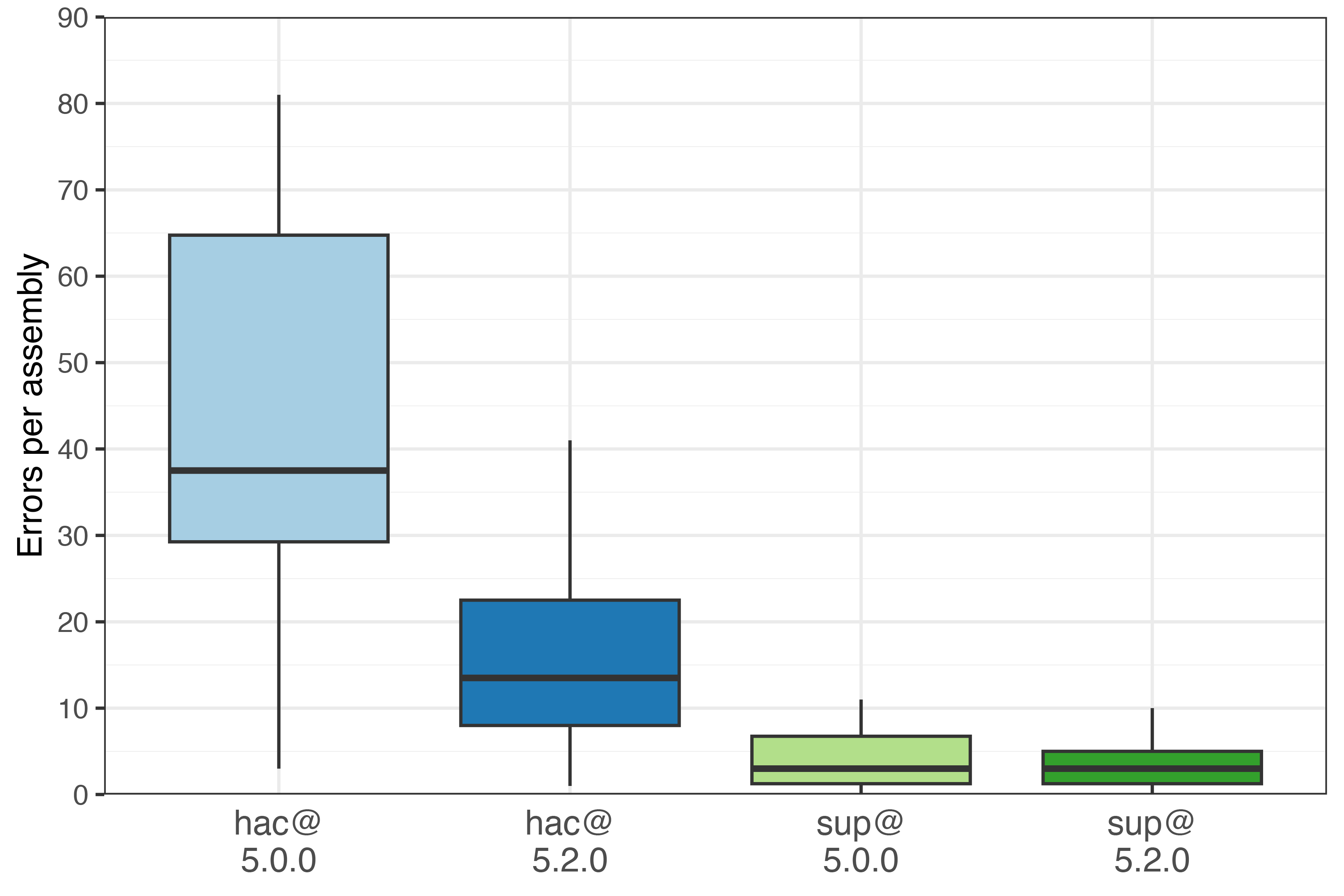
For hac, median errors per assembly dropped from 37.5 with hac@5.0.0 to 13 with hac@5.2.0 – a 65% reduction, despite only a 15% drop in read errors. Very nice! The mean assembly accuracy was Q50.6 for hac@5.0.0 and Q54.5 for hac@5.2.0.
For sup, both sup@5.0.0 and sup@5.2.0 had a median of 3 errors per assembly.4 Still, the newer model improved the error rate for some assemblies, reducing the mean errors per assembly by ~1. The mean assembly accuracy was Q60.4 for sup@5.0.0 and Q61.6 for sup@5.2.0.
Across all 30 sup@5.2.0 assemblies, I counted 99 bp of errors in total. These occurred at 79 loci: 44 were homopolymer-length errors (indels up to 4 bp), 22 were 1-bp substitutions, 12 were 1-bp indels and one was a 2-bp substitution.
Discussion and conclusions
This new release is a big upgrade for hac users: modest read accuracy gains and large improvements in assembly accuracy. The downside is that hac basecalling is now a little bit slower, but I think that trade-off is worth it.
However, I almost exclusively use sup basecalling, so the hac improvements aren’t very important to me. The new sup model did improve assembly accuracy, but not by much. Given that a full year passed since the last model release, I was hoping for a bigger improvement in sup assemblies.
ONT’s accuracy has improved rapidly over the past decade, which has been exciting but also frustrating for users who want stability. In my domain (bacterial whole genome sequencing), this past year has been unusually stable. I appreciate not feeling the need to rebasecall my data every few months, but I also miss the thrill of shrinking error rates. My guess is that ONT has already picked the low-hanging fruit, and the assembly errors that remain are genuinely hard to avoid. I’m still hoping for a day when ONT-only assemblies can be reliably perfect, but for now, short-read polishing is still often needed to clean up the last few errors.
This post looked at Autocycler assembly accuracy without any post-assembly polishing. I noticed that new Dorado polish models were released alongside the new basecalling models, but there’s no updated bacterial model. In a previous post, I found the bacterial model outperformed the move-table-aware sup model for bacterial isolates. So here’s a request for ONT: please train a sup@5.2.0 move-table-aware bacterial model for Dorado polish! I’m optimistic it would be the best option for polishing ONT-only bacterial genome assemblies.
Footnotes
-
I think the basecalling model version (e.g.
hac@v5.0.0vshac@v5.2.0) is more relevant than the Dorado version (0.9.5 vs 1.0.0). I used Dorado v0.9.5 (released a couple months ago) with the 5.0.0 models because I had already done thesup@5.0.0basecalling for the Autocycler preprint. ↩ -
I used what Heng Li calls BLAST identity: matching bases divided by alignment length. That’s column 10 / column 11 from a
minimap2 -cPAF file. ↩ -
These plots show how ONT reads have a bimodal identity distribution: some reads cluster around Q5–Q10, while most peak above Q15. I’ve seen this pattern before (not unique to this run), but I don’t know the cause. If you do, let me know! ↩
-
In case anyone is closely checking my work: the curated assemblies in the Autocycler preprint (which used
sup@5.0.0) had a median of 4 errors per assembly, but I report a median of 3 in this post. That’s because I re-ran the assemblies for this analysis, and full Autocycler runs aren’t completely deterministic. Autocycler itself is deterministic, but some of the input assemblies it relies on are not. ↩
