Medaka vs Dorado polish
For years, Medaka has been the standard tool for polishing genome assemblies with ONT reads. However, with the release of Dorado v0.9.0 in December 2024, ONT introduced a new polishing option: the dorado polish command.
Shifting polishing to Dorado makes sense from a computational perspective. Dorado is written in C++ with libtorch and optimised for both NVIDIA and Apple GPUs, making it fast and efficient. But what really caught my attention was this note in the Dorado README:1
When auto or <basecaller_model> syntax is used and the input is a v5.0.0 dataset, the data will be queried for the presence of move tables and the best polishing model will be selected for the data. Move tables need to be exported during basecalling. If available, this allows for higher polishing accuracy.
Move tables store information about dwell times – how long each base lingers in the pore – which provides additional context for the neural network during polishing.2 Since Dorado polish can use this move-table data, it has the potential to outperform Medaka, which does not use it.
Initially, Dorado polish was not recommended for bacterial genomes,3 but the recent release of Dorado v0.9.1 introduced a bacterial polishing model. In this post, I test Dorado polish on a set of bacterial genomes and compare it to Medaka.
Methods
I used 10 bacterial genomes from a P2 Solo run, each from a different species.4 These genomes had deep ONT sequencing coverage and complementary Illumina reads.
To take advantage of Dorado’s move-table functionality, I first re-basecalled the ONT reads with the --emit-moves option, using the sup@v5.0.0 basecalling model. An exciting side note: Dorado v0.9.1 includes performance optimisations that made basecalling about 50% faster than before!5
Next, I created high-quality reference assemblies using Autocycler, Polypolish and Pypolca. Everything went well, so I assume my reference genomes are error-free (or close to it).
To generate draft assemblies for polishing, I wanted ONT-only genome assemblies with a meaningful number of errors. My existing Autocycler ONT-only assemblies (used for generating the reference assemblies) were too accurate, containing just 21 total errors across all 10 genomes, with some genomes being completely error-free. To better assess polishing performance, I intentionally created lower-quality assemblies by:
- Randomly subsampling the ONT reads to 50× coverage.
- Assembling with Raven.
- Manually correcting any large indel errors.6
This produced 10 draft assemblies containing a total of 270 errors.7
I then polished each genome using five different methods:
- Medaka-bac: Medaka with the
r1041_e82_400bps_bacterial_methylationmodel. This is the model Medaka auto-selects when using the--bacteriaoption and is the recommended Medaka model for native bacterial DNA. - Medaka-sup: Medaka with
r1041_e82_400bps_sup_v5.0.0model. This is the model Medaka auto-selects when not using--bacteria. - Dorado-bac: Dorado with the
dna_r10.4.1_e8.2_400bps_polish_bacterial_methylation_v5.0.0model. This is the model Dorado auto-selects when using the--bacteriaoption and is the recommended Dorado model for native bacterial DNA. This model does not make use of move-table data.8 - Dorado-sup: Dorado with the
dna_r10.4.1_e8.2_400bps_sup@v5.0.0_polish_rlmodel. This is the model Dorado auto-selects when not using--bacteriaand when move-table data is not present. - Dorado-sup-mv: Dorado with the
dna_r10.4.1_e8.2_400bps_sup@v5.0.0_polish_rl_mvmodel. This is the model Dorado auto-selects when not using--bacteriaand when move-table data is present.
Here are some details of each model’s neural network:
| Polishing model | Architecture | Weights |
|---|---|---|
| Medaka-bac | 2-layer bidirectional GRU | 405253 |
| Medaka-sup | 2-layer bidirectional GRU | 405253 |
| Dorado-bac | 2-layer bidirectional GRU | 405253 |
| Dorado-sup | CNN + 4-layer LSTM | 4853501 |
| Dorado-sup-mv | CNN + 4-layer LSTM | 4853565 |
Other notes:
- I used the 50× subsampled ONT reads as input for polishing, not the full ONT read set.
- I gave each tool 32 threads to use, but they actually used far fewer. Dorado in particular spent most of its runtime on a single thread.
- While both Medaka and Dorado support GPU acceleration, I ran all tests on CPU (an AMD EPYC 7742). The smaller models (Medaka-bac, Medaka-sup, Dorado-bac) ran efficiently on CPU, but the larger models (Dorado-sup, Dorado-sup-mv) were slower and would likely benefit from GPU acceleration.
Results
| Polishing method | Total errors |
Overall qscore |
Time (h:m:s) |
RAM (GB) |
|---|---|---|---|---|
| draft assemblies (no polishing) | 270 | Q52.2 | n/a | n/a |
| Medaka-bac | 26 | Q62.4 | 0:02:09 | 7.2 |
| Medaka-sup | 100 | Q56.5 | 0:01:44 | 7.1 |
| Dorado-bac | 25 | Q62.5 | 0:03:30 | 10.3 |
| Dorado-sup | 203 | Q53.4 | 1:00:35 | 46.5 |
| Dorado-sup-mv | 101 | Q56.5 | 1:02:23 | 46.7 |
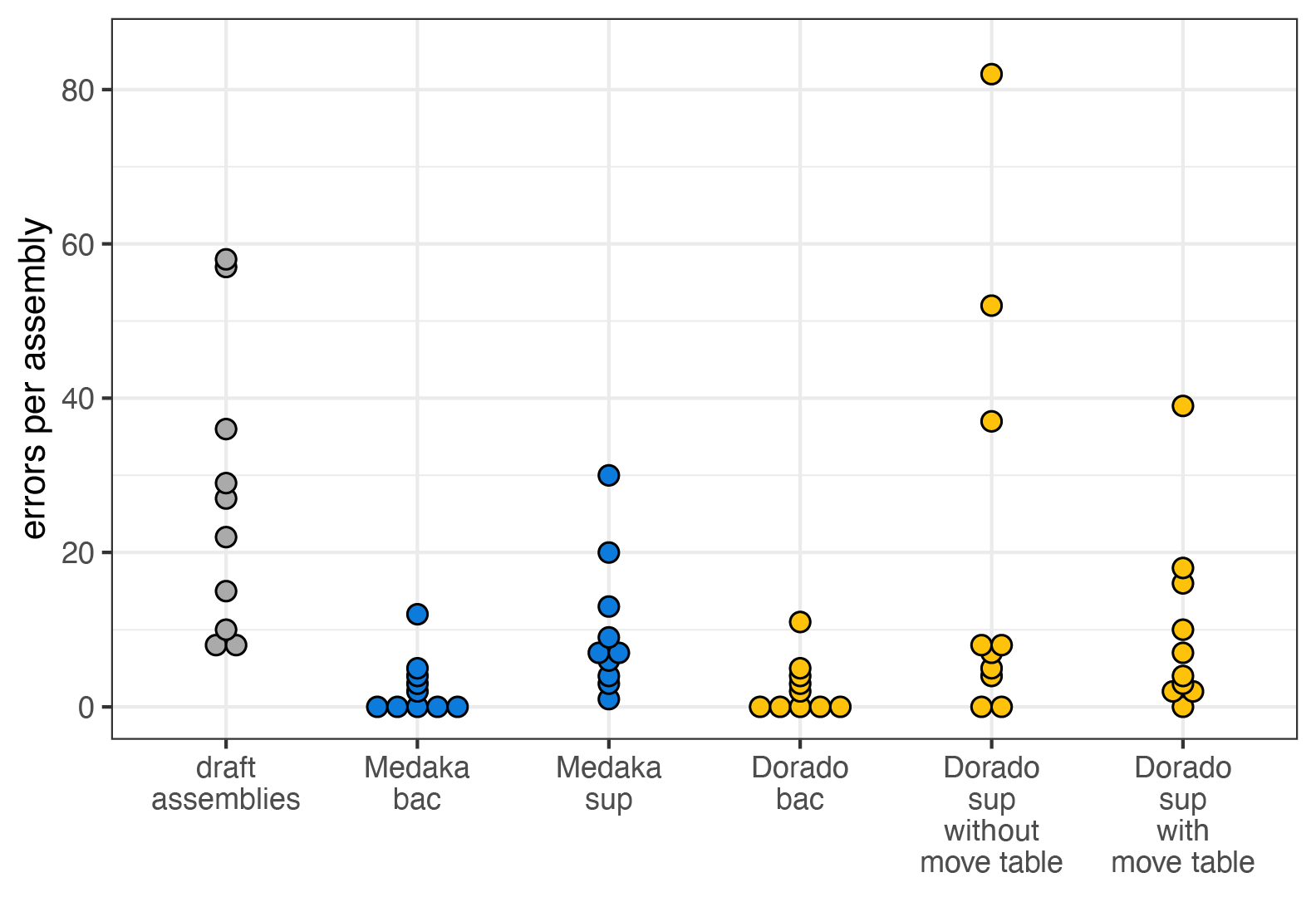
- Total errors: Sum of all errors across the 10 genomes. Per-genome results are in the figure below.
- Overall qscore: Calculated from total errors and total genome size (44.9 Mbp).
- Time and RAM: Median values across the 10 genomes.
Discussion
Medaka-bac and Dorado-bac were the best-performing polishers, each reducing errors by about 10-fold (Q52 → Q62). Given that these are ONT’s recommended models for bacterial genome polishing, this result was expected. However, what stood out was that Medaka-bac and Dorado-bac produced nearly identical results – only a 1 bp difference across all 10 genomes. This led me to suspect that these models were not just similar in architecture but also identical in their trained weights. I confirmed this by checking with PyTorch, and yes, Medaka-bac and Dorado-bac share the exact same weights. At the time of writing, this means Medaka and Dorado are interchangeable for bacterial genome polishing. However, it seems likely that ONT will transition to Dorado as the recommended tool, which may lead to Medaka becoming deprecated in the future.9
The Medaka-sup model performed worse than Medaka-bac, which isn’t surprising – it was presumably trained on data with less bacterial diversity and more non-bacterial reads. The Dorado-sup and Dorado-sup-mv models also did poorly. This aligns with ONT’s notes stating that these models are optimised for human genomes.10
Even though Dorado-sup and Dorado-sup-mv performed poorly on bacterial genomes, the move-table-aware model (Dorado-sup-mv) outperformed its non-move-table counterpart (Dorado-sup). This suggests that move-table data is indeed beneficial for polishing. Also, these models have a larger and more sophisticated neural network architecture than Dorado-bac. This raises an interesting question: What if ONT trained a bacterial-specific move-table-aware model with this bigger architecture? This hypothetical Dorado-bac-mv model could potentially outperform Dorado-bac. If and when such a model is released, I’ll test it out in a follow-up post.
One more interesting feature of Dorado polish: it has a --qualities option which makes Dorado output the polished genome in FASTQ format. This has interesting implications, but after drafting a section on it, I realised the topic is big enough for its own blog post, so stay tuned for that!
Footnotes
-
Minor typos were corrected for clarity. ↩
-
For more on move tables, see this explanation in Dorado’s README and this technical document from squigualiser. ↩
-
See the Dorado v0.9.0 release notes and Mike Vella’s Bluesky post. ↩
-
The genomes are Enterobacter hormaechei, Enterobacter kobei, Escherichia coli, Klebsiella planticola, Klebsiella pneumoniae, Listeria innocua, Listeria monocytogenes, Listeria seeligeri, Providencia rettgeri and Shigella flexneri. And yes, I know that Shigella is technically E. coli, so that’s really only nine unique species. ↩
-
When I last tested Dorado’s speed on an NVIDIA A100 GPU, it basecalled at 6.76e+06 samples/sec. This time, it basecalled at 1.04e+07 samples/sec. Also see the Feb 2025 update in my Spring OnION post. ↩
-
I manually corrected large indels because polishing tools are expected to fix small errors but not necessarily large structural errors. After corrections, all remaining errors in my draft assemblies were either substitutions or indels of ≤10 bp. ↩
-
Errors were counted per base. For example, a 5-bp deletion counts as 5 errors. If instead each indel were counted as a single error regardless of size, the total count across all 10 draft assemblies would be 174 errors. ↩
-
I initially tested Dorado-bac both with and without move-table data, but the results were identical. Later, I confirmed that the Dorado-bac model is identical to the Medaka-bac model, meaning it does not use move-table data. ↩
-
See Chris Wright’s comment here: github.com/nanoporetech/medaka/issues/547 ↩
-
The Dorado v0.9.0 release notes (before the bacterial model was added) state that Dorado polish ‘is optimised for refining draft assemblies of human genomes.’ ↩
