FASTQ assemblies with Dorado polish
This post builds on my previous one about using Dorado for genome polishing. Its key takeaway: Dorado polish and Medaka use the same bacterial polishing model, making them effectively interchangeable for bacterial genomes (at least at the time of writing).
While writing that post, I noticed something interesting in the Dorado docs: using the --qualities option outputs the polished assembly in FASTQ format. I initially thought this was unique to Dorado, but I checked Medaka, and it actually has a -q option that does the same thing, so this feature has been hiding under my nose for a while now.1
The key difference between FASTA and FASTQ is that FASTQ includes per-base quality scores, encoded as ASCII characters.2 In a well-calibrated FASTQ, these scores indicate the absolute probability of an error, but if not well-calibrated they can still indicate relative error likelihood.
FASTQ is commonly used for reads, while assemblies typically use FASTA. But assemblies can use FASTQ format, and it could be useful in downstream analyses. For example, when calling variants from an assembly, errors can create false positives.3 FASTQ quality scores could help by allowing one to mask low-confidence bases in the assembly.
So what do Dorado polish FASTQs look like? How well do low qscores correlate with assembly errors? Are the scores well calibrated? In this post, I take an initial look using the same genomes from my previous post, focusing on the five that still had errors after Dorado-bac polishing.
Calibration
To assess how well the FASTQ quality scores are calibrated, I calculated the expected number of errors per genome using \(\sum 10^{-q/10}\) summing over all genomic positions.
| Genome | Actual errors | Expected errors |
|---|---|---|
| Shigella flexneri | 5 | 58.4 |
| Klebsiella pneumoniae | 3 | 19.4 |
| Providencia rettgeri | 2 | 16.9 |
| Enterobacter kobei | 11 | 31.7 |
| Escherichia coli | 4 | 23.5 |
While not perfectly calibrated, the predictions are within an order of magnitude or so – better than I expected!
Qscore plots at error locations
Across the five genomes, there were 25 bp of errors at 17 loci. Each locus is plotted below, with qscores on the y axes. The x axes display the assembled sequence (top) and ground truth (bottom, sometimes with extra bases squeezed in when the assembly had a deletion). Red bars mark error positions, and for deletion errors I used red on both sides of the deletion.
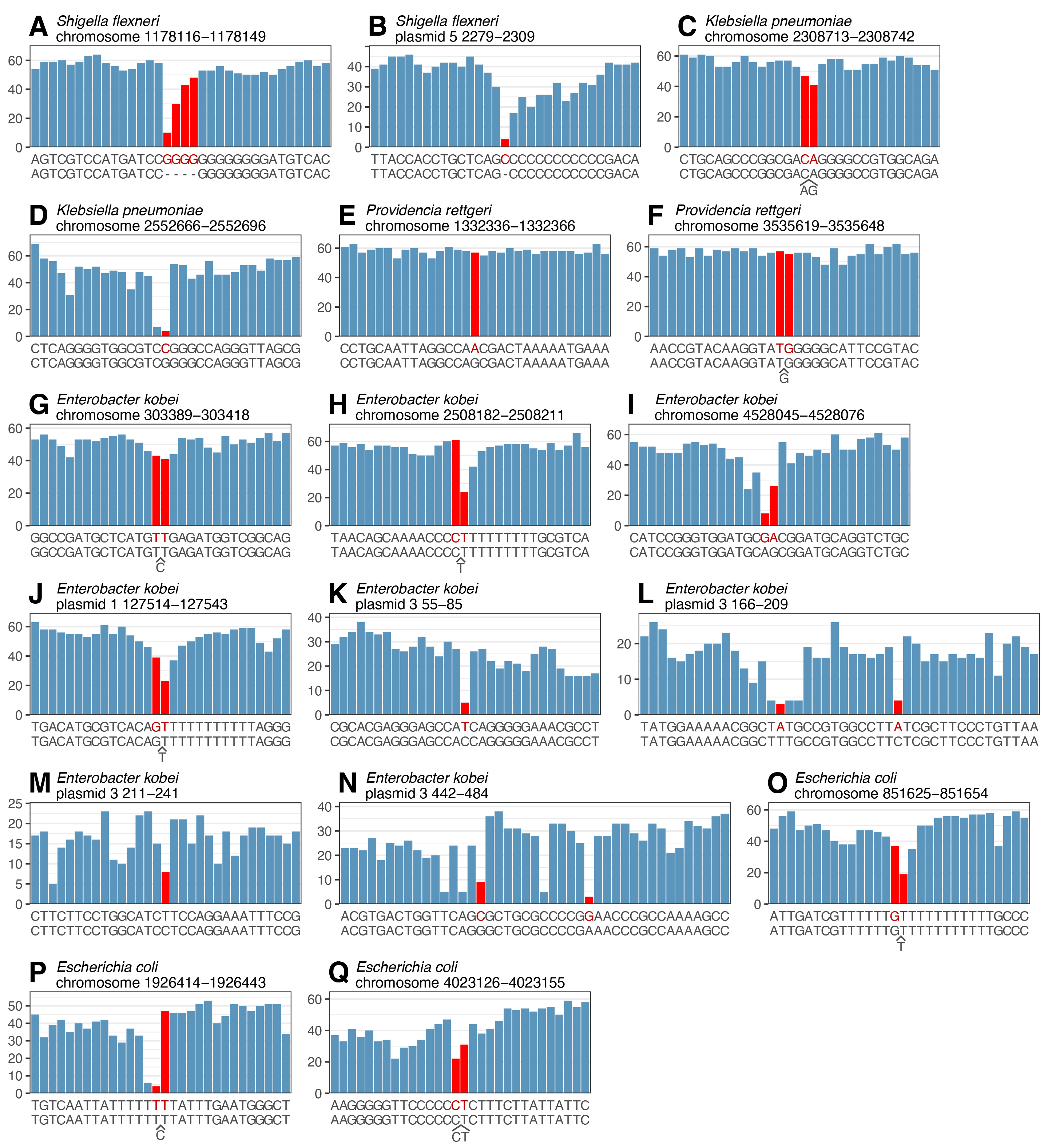
As you can see, positions with or near errors often have a much lower qscore than their neighbours. This is excellent, as it means the FASTQ scores indeed provide a useful indicator of base reliability.
However, there are exceptions. In particular, E and F lacked qscore drops. On closer inspection, the ONT reads at these sites were very clear (no heterogeneity) but differed from the Illumina reads, suggesting these might not be errors at all.4 Since my ONT and Illumina reads came from separate DNA extractions, biological differences between the read sets are possible. Even discounting E and F, some errors showed only subtle qscore drops. For example, C and G appear to be real errors, yet their qscores remain above 40.
Discussion
These results just scratch the surface, but they show that Dorado-polish qscores, while imperfect, are potentially quite useful. Using my previous example of calling variants from an ONT assembly, one could reduce false positives by masking low-quality bases (perhaps with a local threshold, e.g. masking bases with a qscore significantly lower than their neighbours). The ideal strategy would balance sensitivity and specificity based on the application.
FASTQ assemblies could also improve genome polishing. I’ve spent a lot of time trying to make short-read polishing more reliable,5 but false-positive corrections can still occur.6 Dorado-polish qscores could help decide which changes to accept. For example, E and F were short-read polishing corrections in the Providencia genome, but their high Dorado-polish qscores could have convinced me to reject those changes.
Even if FASTQ output is a niche feature, I’m glad Dorado includes it. FASTQ assemblies provide a useful way to assess base-level accuracy, even when qscore calibration isn’t perfect. I think we should embrace FASTQ as an assembly format, especially for workflows that could benefit from identifying unreliable bases.
Footnotes
-
The
-qoption was added tomedaka_consensusin v1.8.1 (June 2023), but it’s undocumented in the Medaka README, which is probably why I missed it. ↩ -
This is usually done with this formula:
ascii - 33 = qscore. So the scale starts with the!character (ascii = 33, qscore = 0) and could potentially go up to the~character (ascii = 126, qscore = 93), but the highest value I saw in a Dorado polish FASTQ wasg(ascii = 103, qscore = 70). ↩ -
I’m about to release a preprint on this very topic – stay tuned! ↩
-
This means the error totals in my previous post are slightly off. I reported the Dorado-bac assemblies as having 25 total errors, but I now think 23 total errors is more accurate. ↩
-
George Bouras and I published this paper last year with a lot of relevant info: How low can you go? Short-read polishing of Oxford Nanopore bacterial genome assemblies. ↩
-
This is especially true when there are genuine biological differences between the short-read and long-read sets, as is likely the case for the Providencia genome in this post. I hate hybrid read sets made from separate DNA extractions! ↩
