HERRO read correction – part 2
In my last post, I explored HERRO’s ability to correct simulated ONT reads using genomes with a 5.2 kbp repeat. Each genome contained one instance of the repeat with a single-bp variant in the middle, and I assessed how well HERRO maintained that variant during its read correction. Importantly, the read length in that test was ~10 kbp, considerably longer than the repeat length. In this post, I examine the tougher scenario where the repeat is longer than the read length.
Here is a plot of the results from my last test, where ‘HERRO success rate’ means how often the corrected reads that span the variant had the correct variant base:
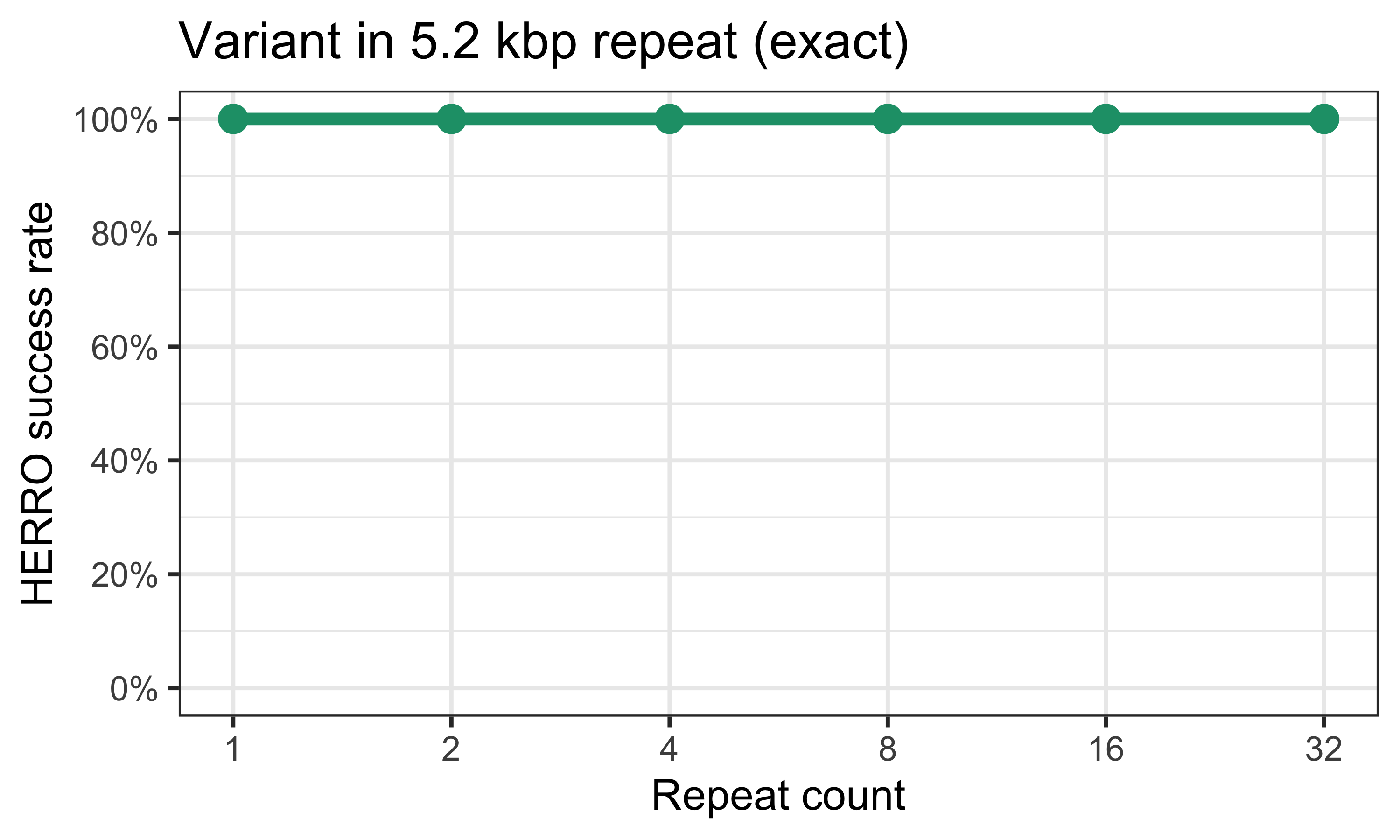
HERRO did so well in that test that the plot isn’t very interesting, but I include it here for comparison with the results below.
Long exact repeat with 1 SNV
For this test, I started with the six artificial genomes used in the previous post, containing 1, 2, 4, 8, 16 and 32 copies of the 5.2 kbp rRNA operon. I then replaced each rRNA operon copy with a 46.3 kbp prophage1, making the repeat almost nine times larger. As before, I introduced a single variant in the middle of one repeat copy.
Now, the read length (~10 kbp) is considerably shorter than the repeat length. This could make HERRO’s job considerably more difficult, as all-vs-all read alignments will no longer be able to easily distinguish different repeat copies.
Here’s a plot of how well HERRO performed with this much longer repeat:2
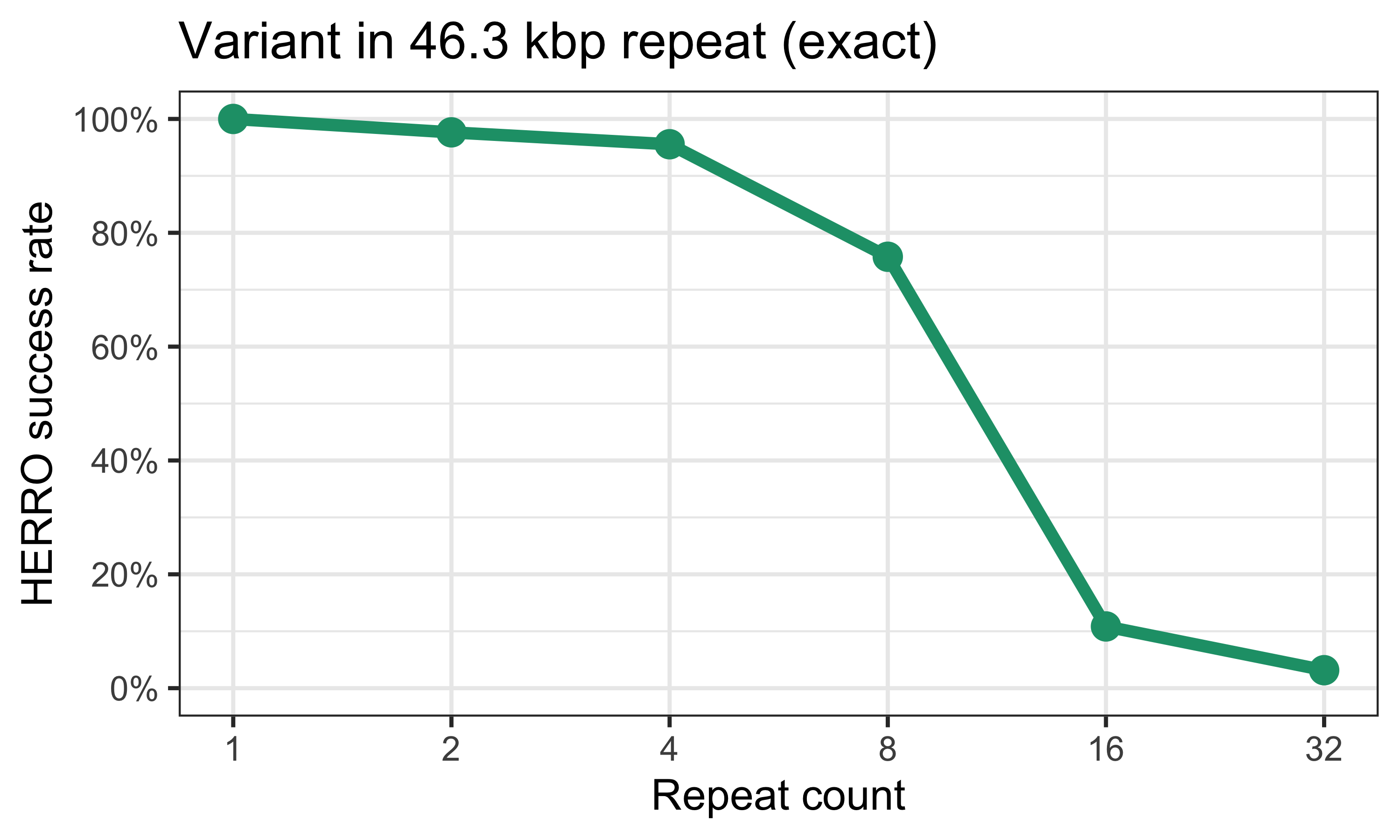
At lower repeat copy numbers, HERRO usually got the variant base correct, but it started to falter when the repeat copy number reached eight and above.
99.9% identity repeat
Next, I wanted to see if HERRO could do better if the repeat wasn’t quite exact. For the instance containing the variant, I introduced additional variants at 1000 bp intervals, resulting in 47 total variants. This made the variant-containing repeat copy have ~99.9% identity to the other copies.
In theory, this should provide HERRO with enough information to get the variant base correct, as my ~10 kbp reads are sufficient to phase these variants. Here are the results:3
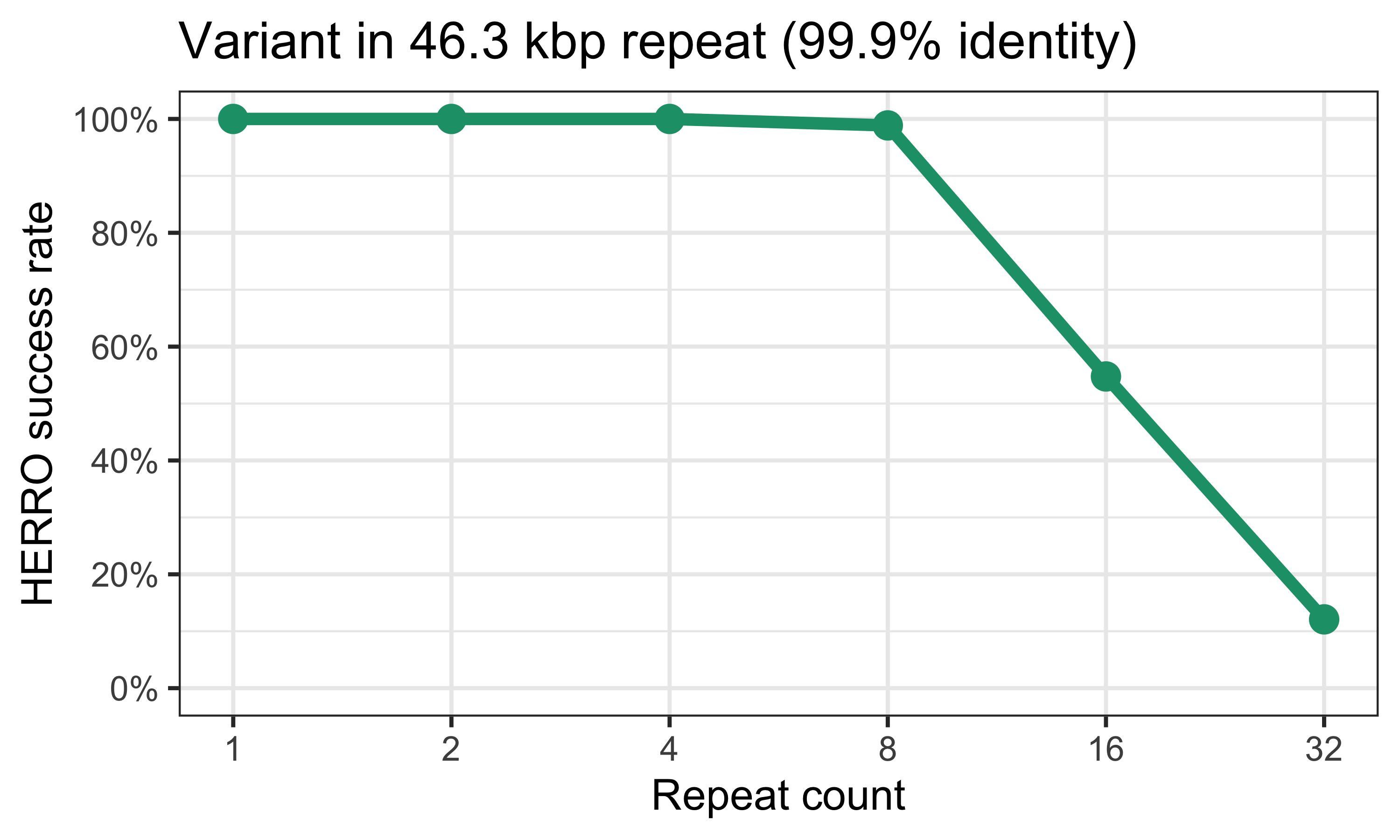
HERRO did better, usually getting the variant base correct up to the 8-copy genome. But it still faltered with the 16-copy and 32-copy genomes.
99% identity repeat
Finally, I made it even easier for HERRO by introducing additional variants at 100 bp intervals, resulting in the variant-containing repeat copy having ~99% identity to the other copies.
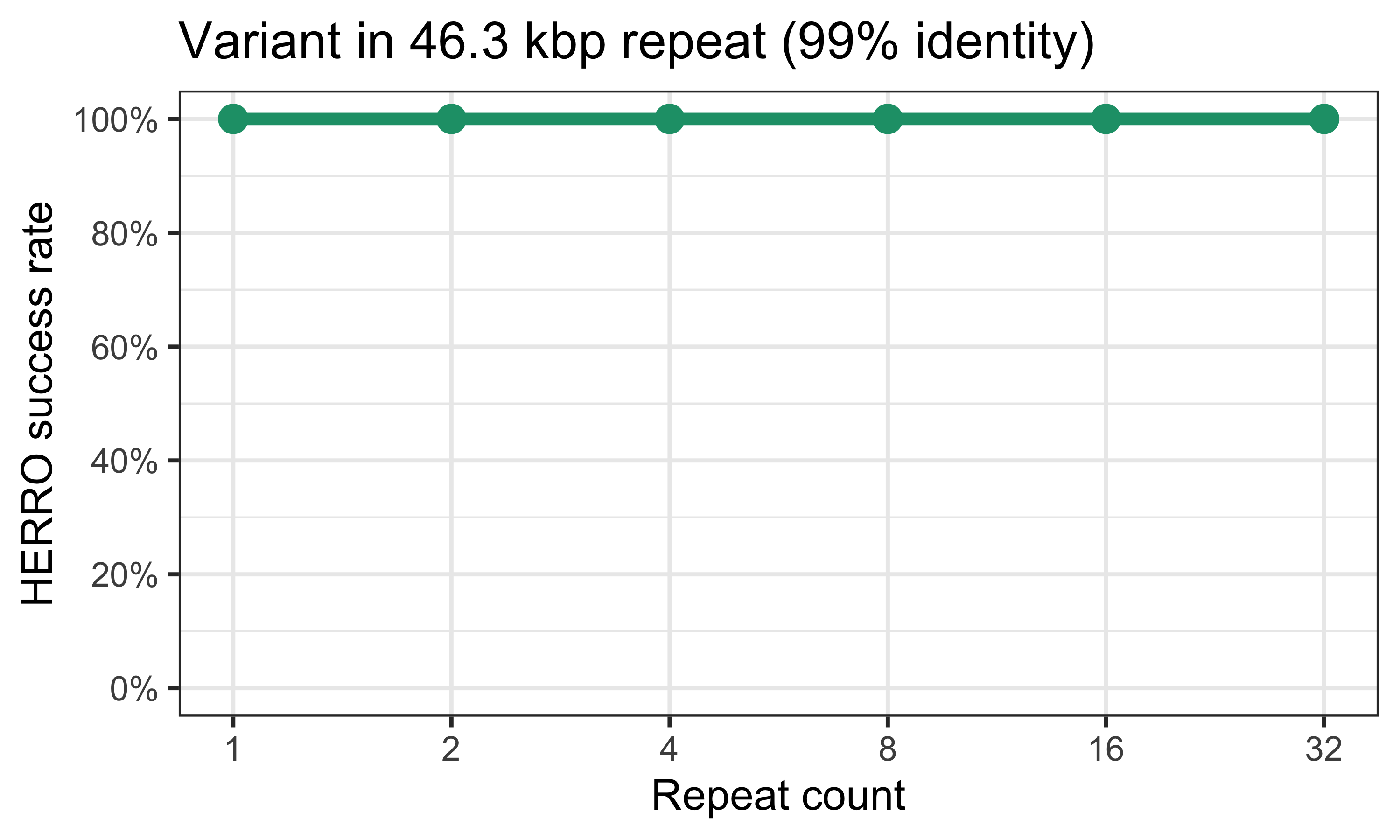
Now HERRO was able to correctly maintain the variant, even in the 32-copy genome.
Discussion and conclusions
While bacterial genomes often contain multiple copies of a prophage, I’ve never seen 32 in a single genome, so my tests in this post were unrealistically challenging. Even so, HERRO did well!
Based on my previous post and this one, here are some final thoughts:
- HERRO works great when read length > repeat length, even with high-copy-number repeats.
- When read length < repeat length, it still performs well with low-copy-number repeats.
- To handle high-copy-number repeats when read length < repeat length, the repeats need some variation to help HERRO distinguish different copies. 99% identity seems to suffice.
Overall, I’m very pleased with HERRO’s performance. Initially, I worried that it might erase minority variants from repeats, but I now trust that it will not, at least in most realistic scenarios for a bacterial genome.
As I mentioned in the first post, these tests were far from comprehensive. I only used simulated reads and synthetic genome sequences, and I did not assess whether HERRO-corrected reads assemble better than uncorrected reads. However, the encouraging results from these initial tests motivate me to try HERRO more, and I will experiment with real read sets in the future.
Footnotes
-
I got this prophage sequence from MGAS6180, found in the supplementary data of this paper. ↩
-
In my first post, I also assembled the HERRO-corrected reads with Flye and Unicycler assemblies. This time I didn’t assemble the reads, because when repeats are longer than reads, I don’t expect assemblies to yield a complete genome sequence. ↩
-
Even though my repeat now has many variants, for simplicity I only assessed HERRO’s accuracy on the same middle-of-the-repeat variant as before. ↩
