Benchmark update: metaMDBG and Myloasm
New assembler releases
Our manuscript describing Autocycler was recently published:1
Wick RR, Howden BP, Stinear TP (2025). Autocycler: long-read consensus assembly for bacterial genomes. Bioinformatics. doi:10.1093/bioinformatics/btaf474.
But its benchmarking is already out of date! Since I ran the analyses for the paper, two long-read assemblers have had new releases: metaMDBG v1.2 and Myloasm v0.2.0. Both came with claims that caught my eye (‘improved assembly quality’ for metaMDBG and ‘cleaner contig outputs with better polishing’ for Myloasm), and both tools are still young (especially Myloasm). I therefore decided to rerun these new versions through the same benchmarking pipeline I used in the Autocycler paper.2
Updated results
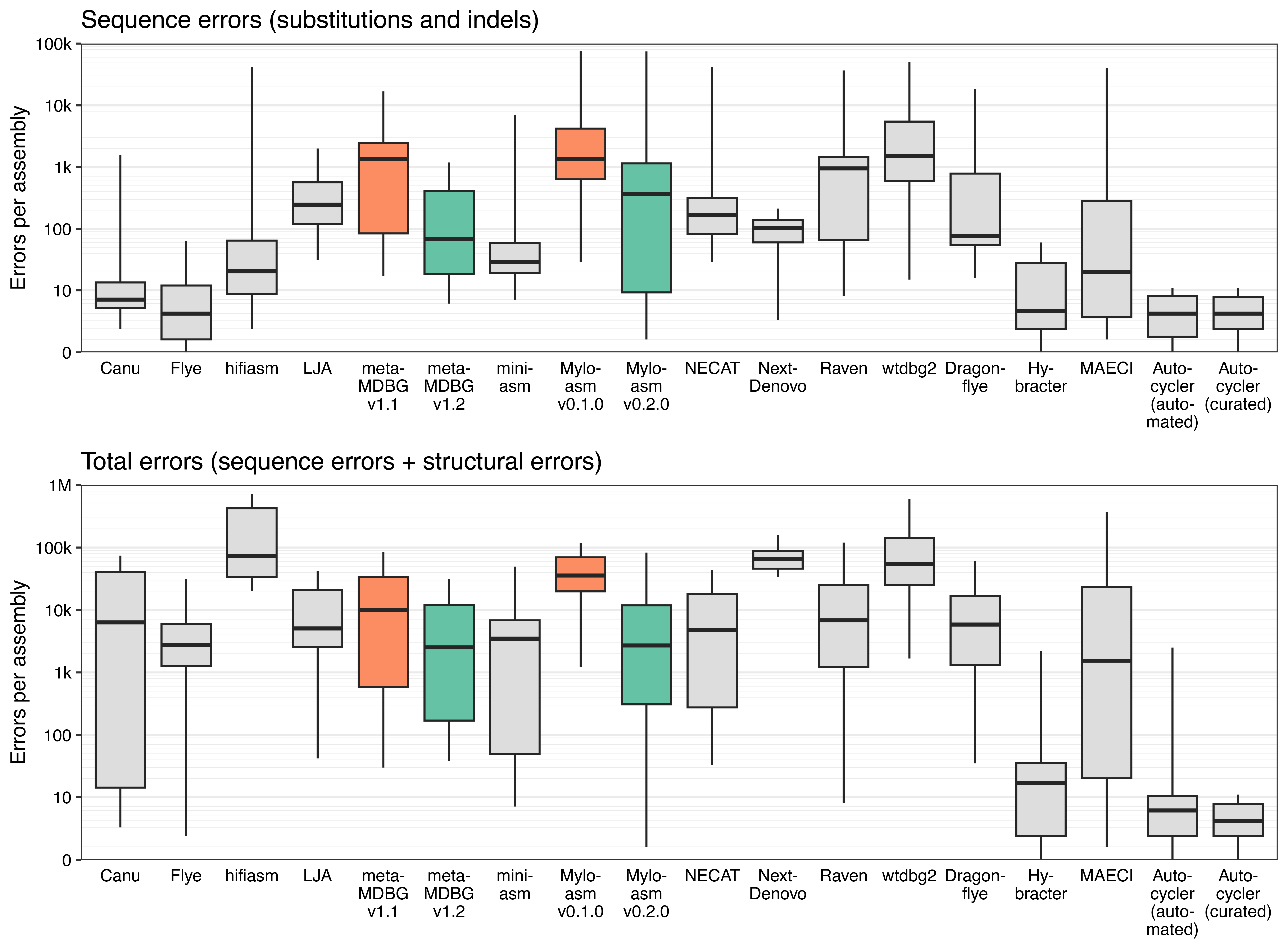
Below is an updated version of Figure 2 from the Autocycler paper. Error counts are shown on the y-axes (pseudo-log transformed, lower is better). The original metaMDBG and Myloasm versions (from the paper) are orange, the new versions are green and everything else (less relevant here) is grey.
I also updated the relevant supplementary figures using the same old-orange new-green colour scheme:
- Figure S1: detailed results by error type
- Figure S2: Inspector results
- Figure S3: CRAQ results
- Figure S4: BUSCO results
- Figure S5: runtime and memory usage
Discussion
Both metaMDBG and Myloasm showed clear improvements in accuracy with their latest releases: fewer sequence errors (substitutions and indels) and fewer total structural errors.3 I was particularly impressed by the best cases for Myloasm v0.2.0 – a couple of the Listeria innocua assemblies had only one single-bp error, better than any other single-tool assembler.
When I run Autocycler, I usually use this Bash script to automate the process. Autocycler benefits from a diverse set of input assemblers, but I had previously left out Myloasm because v0.1.0 had relatively high error rates. These new results, along with positive reports from a colleague4, convinced me to add Myloasm to the pipeline.
It’s worth noting that both metaMDBG and Myloasm were developed as metagenome assemblers, but I’m using them here to assemble isolate genomes. As my results show, metagenome assemblers can work quite well on isolates! However, they can be more likely to leave low-depth contigs in the assembly. In metagenomes this is desirable, since there are often many low-abundance organisms. But for isolates, low-depth contigs usually indicate contamination.5 For these tests, I ran the assemblies via Autocycler helper using --min_depth_rel 0.1 to remove contigs below 10% chromosomal depth, and I recommend others do the same when applying these assemblers to isolates.
Footnotes
-
At the time of writing, the paper is reviewed and accepted but still an unproofed advance article. ↩
-
For the full methods and results, see the Autocycler paper GitHub repo. ↩
-
The only metric that got worse with the new versions is ‘missing bases’, but this was balanced by improvements in the ‘extra bases’ metric (see Figure S1). ↩
-
Michael Hall had one tricky genome where metaMDBG and Myloasm were the only two assemblers which could successfully assemble the chromosome. ↩
-
A common cause would be cross-barcode contamination. In multiplexed ONT runs, some reads can ‘leak’ into other barcodes, and if the source is sufficiently high depth (e.g. a high-copy-number plasmid), the contamination can sometimes reach assemblable levels in wrong barcodes. ↩
