Cross-sample homopolymer polishing with Pypolca
Cryptosporidium assembly
I was recently working with Torsten Seemann and our colleagues at the Centre for Pathogen Genomics to assemble a Cryptosporidium hominis genome from ONT reads. It’s small for a eukaryote at only 9.2 Mbp, has low GC content (~30%) and, importantly for this post, has lots of long A/T homopolymers (~100 of them ≥20 bp). The ONT sequencing was deep, and the assembly went smoothly: T2T for all eight chromosomes.1 However, I expect it contains homopolymer-length errors – while R10.4.1 is better than R9.4.1, I’m still suspicious of homopolymers >10 bp.
To illustrate the problem, I took a long homopolymer from the Crypto genome and used squigulator to simulate the raw ONT signal. For long stretches of a single base, the signal flatlines, making it difficult for the basecaller to determine the precise homopolymer length:
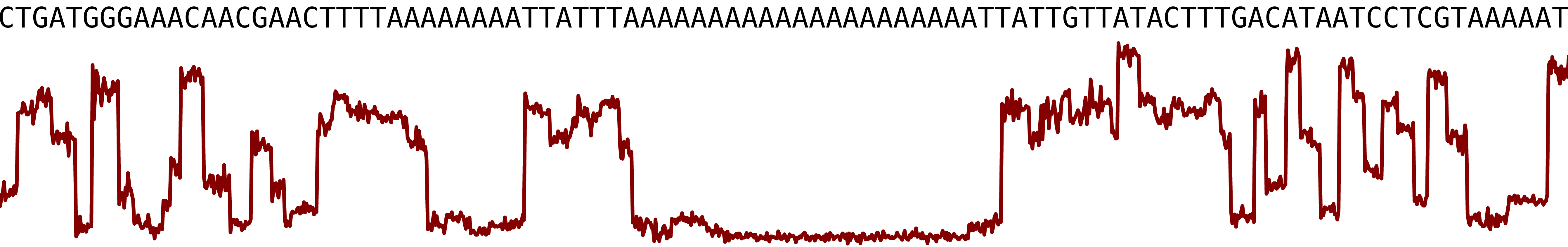
Normally, I’d fix homopolymer lengths (and any other lingering errors) with short reads from the same isolate using Polypolish and Pypolca. But we didn’t have short reads for this genome, so I wondered: could I correct homopolymers using short reads from a closely related isolate instead?2 I’ll call this cross-sample homopolymer polishing.
Homopolymer-only polishing
So I decided to add a homopolymer-only polishing feature to Pypolca. When used, Pypolca will ignore all changes except for length adjustments in homopolymers above a threshold. George Bouras kindly merged my pull request and released it in v0.4.0, so this feature is now available. Documentation is here.
To try it, I downloaded all NCBI assemblies of Cryptosporidium and identified the closest relative to my genome using Mash. I then downloaded Illumina reads for that sample3 and polished with:
pypolca run -a draft.fasta -1 SRR1557959_1.fastq.gz -2 SRR1557959_2.fastq.gz -t 16 --homopolymers 6
This changed the length of 343 homopolymers:4
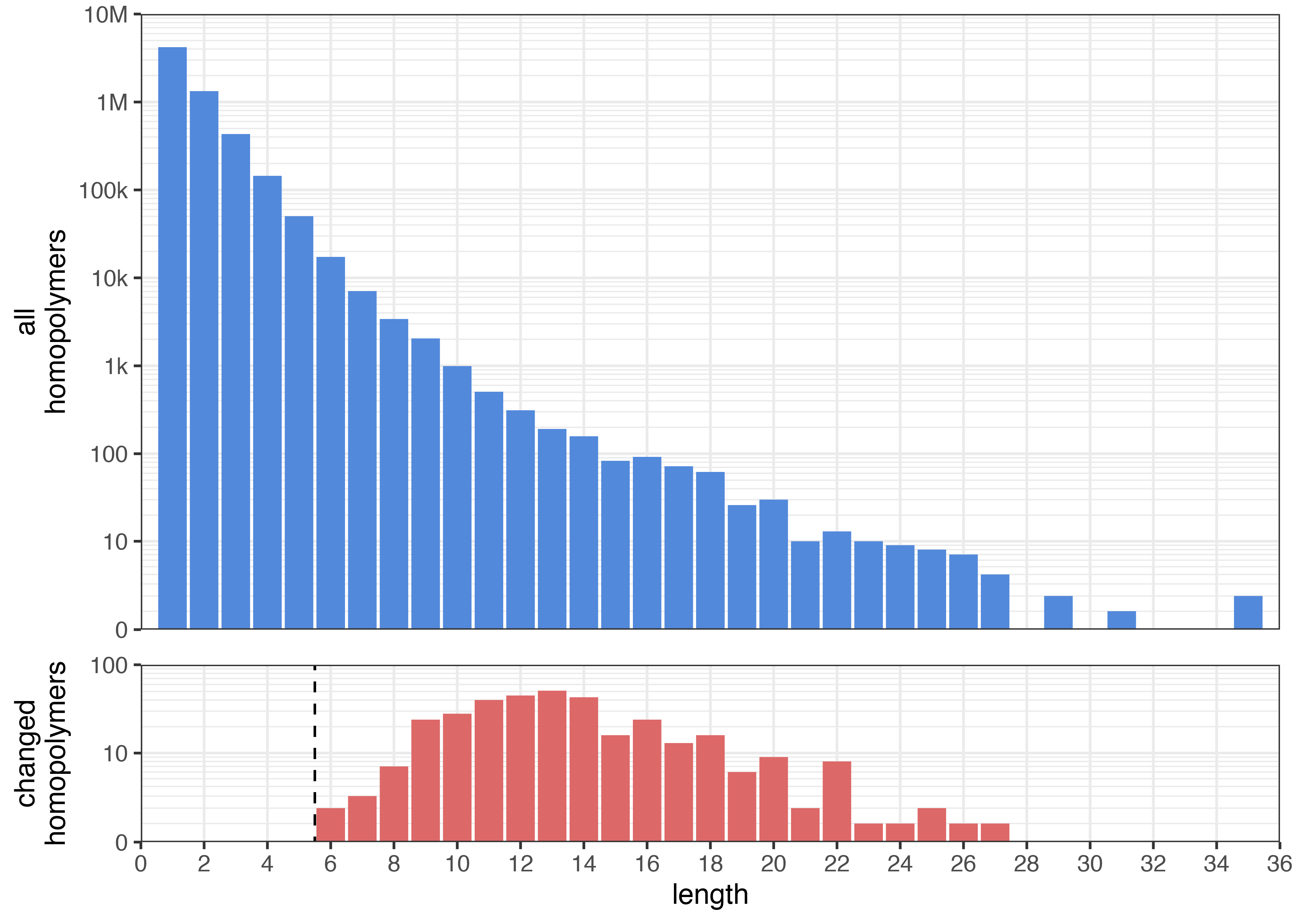
The top plot shows the distribution of all homopolymers in the genome.5 The bottom plot shows homopolymers whose length changed. Both plots use a pseudo-log y axis. The dashed line marks the --homopolymers 6 threshold, i.e. shorter homopolymers were not allowed to change.
The results match my expectations: very few shorter homopolymers were altered (e.g. 2/17297 6-mers and 3/7070 7-mers) where ONT is reliable, but about one-quarter of longer homopolymers changed (e.g. 6/26 19-mers and 9/30 20-mers) where ONT struggles.
I also annotated the genome before and after homopolymer-only polishing with Companion, and it seemed to improve the annotation: gene count increased (4069 → 4076), pseudogene count decreased (41 → 36) and the fraction of bases annotated increased (76.968% → 76.979%).
Discussion
Cross-sample homopolymer polishing looks promising, but it depends on several assumptions:
- ONT-only assemblies have length errors in long homopolymers.
- Illumina reads handle long homopolymers better than ONT reads.
- Closely related genomes usually share the same homopolymer lengths.
- Homopolymer lengths are biologically consistent (i.e. little true variation).
I ordered these by my confidence. Assumption 1 seems safe. I think assumption 2 is usually true, though short reads can struggle in extreme GC contexts.6 I’m less sure about assumptions 3 and 4, which probably depend on the organism – what holds in Crypto may not in other taxa.
So while Pypolca now supports cross-sample homopolymer-only polishing, treat the biological validity as experimental. If the assumptions hold, it may work very well. For our Crypto genome… I’m not yet sure. We’ll be sequencing our isolate with Illumina to get a firmer answer.
Footnotes
-
This was my first time trying Autocycler on a eukaryote genome. It worked well, except for the telomeres at contig ends, which I had to manually repair. See the Autocycler graph here. ↩
-
I couldn’t find an existing tool that does exactly this. This anvi’o script is similar, but it uses a reference genome (not reads) to set homopolymer lengths. Homopolish is also similar, but it can make non-homopolymer changes (see this issue). ↩
-
The source of these reads: Comparative genomic analysis reveals occurrence of genetic recombination in virulent Cryptosporidium hominis subtypes and telomeric gene duplications in Cryptosporidium parvum. ↩
-
For comparison, Pypolca with default settings (not homopolymer-only) made >1600 changes, most of which were not in homopolymers. Some may have been genuine fixes, but I suspect most are biological differences between our genome and the downloaded reads. ↩
-
These homopolymers are much longer than I’m used to seeing in bacteria! As a comparison, I generated random sequences with the same length and GC content, and their longest homopolymer was typically around 15-16 bp. ↩
-
Because homopolymers are pure
A/TorG/C, they skew local GC. In Crypto, all long homopolymers areA/T, which combined with an already low average GC (~30%), means they often fall in very low-GC regions (<20%). Some Illumina preps (e.g. Nextera XT) do not do well with this. ↩
