A first look at CycloneSEQ data
MGI released their CycloneSEQ-WT02 nanopore sequencer this year, which bears a notable resemblance to ONT’s GridION.1 See this preprint for details on CycloneSEQ.
As a long-time user of ONT sequencing, I was curious how CycloneSEQ data compares. In this post, I use publicly available bacterial data to quantify CycloneSEQ’s read-level and consensus-level accuracy and compare it to ONT data.
Data
This preprint used both CycloneSEQ long reads and DNBSEQ short reads to assemble an ATCC type strain (Akkermansia) and 10 other bacterial genomes. It mostly focused on the accuracy of Unicycler hybrid assemblies, where accuracy is primarily determined by the short reads. CycloneSEQ-only assembly accuracy was only mentioned briefly, e.g. for their Akkermansia type strain assembly, where they reported 141 errors per 100 kbp, equivalent to 99.86% accuracy (Q28.5). The preprint also doesn’t focus on read-level accuracy – they show qscores in Figure 1 (average of Q14.4), but these seem to be from the FASTQ files, not the actual read accuracy as measured against a ground-truth reference.
The reads are available on the China National GeneBank DataBase, but downloading them was challenging2, so I only tested one of their genomes: E. coli AM114-O-1. The sequencing was very deep: over 500× short-read depth and 1200× long-read depth.
For an ONT comparison, I used an E. coli genome we recently sequenced with both ONT and Illumina. This wasn’t as deep as the CycloneSEQ data, but still more than enough: over 200× short-read depth and 200× long-read depth. I basecalled the ONT reads with Dorado v0.8.3 using the v5.0.0 models at each accuracy level: fast, hac and sup. Our E. coli is a different strain from the CycloneSEQ E. coli3, so the comparison isn’t perfect but good enough for a first impression.
Methods
I had four datasets to assemble: CycloneSEQ, ONT-fast, ONT-hac and ONT-sup, each with complementary short reads. For short-read QC, I ran fastp with default settings. For long-read QC, I only excluded reads <1 kbp in length.4 The ONT data was part of a barcoded run, so the demultiplexing process also served as a form of QC.5
I assembled each long-read set with Autocycler v0.1.16 to make a long-read-only assembly. I then polished with Polypolish v0.6.0 and Pypolca v0.3.17 to make a ground-truth assembly.
To quantify consensus-level accuracy, I aligned the long-read-only assembly to the ground-truth assembly, counting base differences.8 For read-level accuracy, I aligned the long reads to the ground-truth assembly and calculated accuracy for alignments ≥10 kbp.
Results
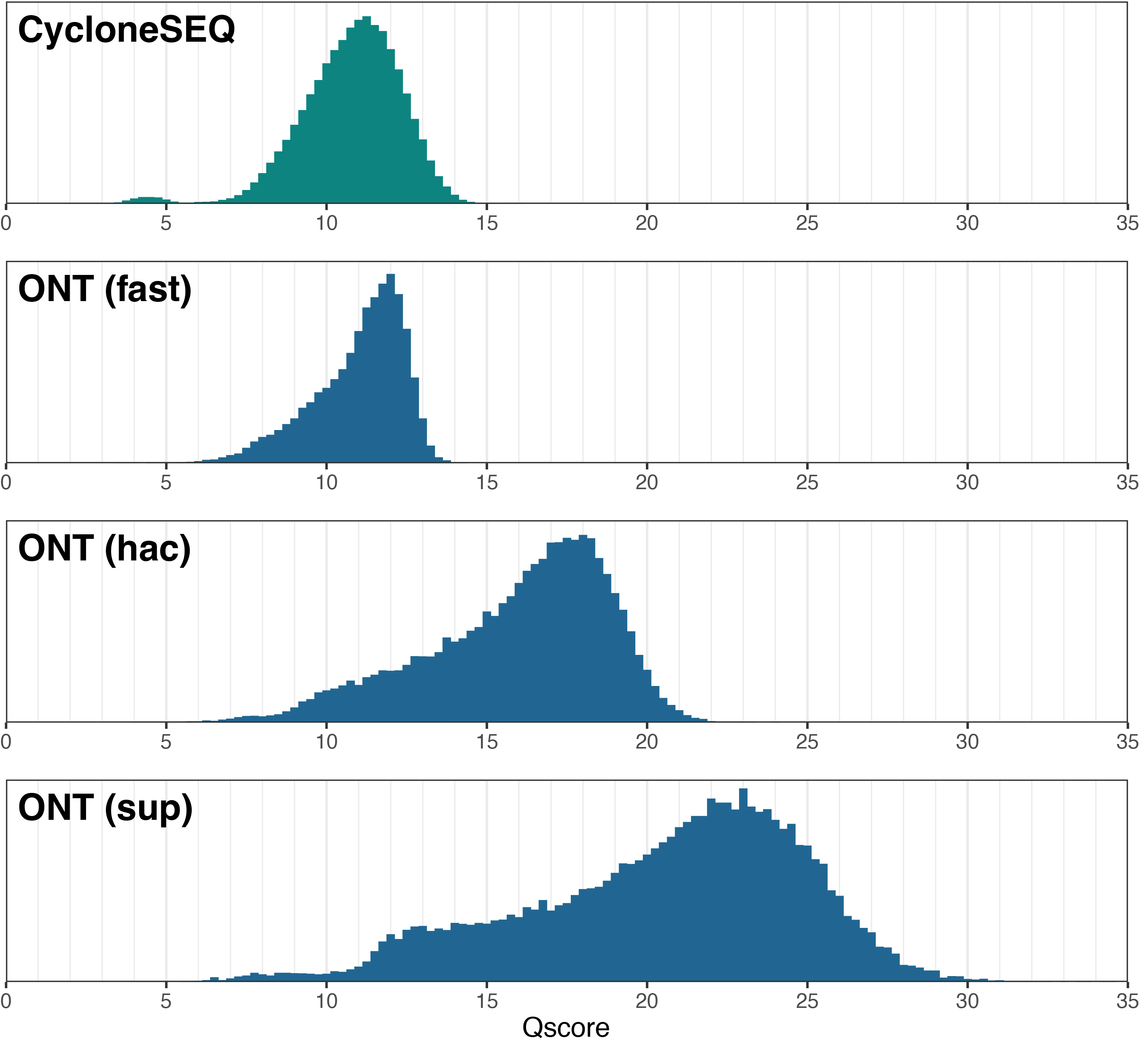
The accuracy metrics are summarised below, with read-level results shown as averages9:
| Genome | Consensus | Read – mean | Read – median | Read – mode |
|---|---|---|---|---|
| CycloneSEQ | 1562 errors (Q35.1) | 90.9% (Q10.7) | 91.9% (Q10.9) | 93.2% (Q11.1) |
| ONT-fast | 2317 errors (Q33.6) | 91.5% (Q11.0) | 92.6% (Q11.3) | 93.8% (Q12.1) |
| ONT-hac | 13 errors (Q56.1) | 96.7% (Q15.9) | 97.8% (Q16.5) | 98.5% (Q17.7) |
| ONT-sup | 2 errors (Q64.3) | 98.4% (Q20.5) | 99.3% (Q21.3) | 99.6% (Q23.1) |
Below are histograms showing read-level accuracy distributions:
Discussion
The preprint that provided the CycloneSEQ data reported read-level accuracy of Q14.4, but my analysis found a much lower value of Q11.1 (modal), probably because I used the ground-truth assembly (not FASTQ scores) to quantify accuracy. For consensus-level accuracy, the preprint reported Q28.5 for their Akkermansia genome, while my analysis of their E. coli genome did better at Q35.1. This could be because the E. coli was an ‘easier’ genome than the Akkermansia or because my assembly method (Autocycler) was more robust. Both the preprint’s results and my analysis found accuracy levels lower than CycloneSEQ’s advertised accuracy of 97% (Q15.2) for reads and 99.99% (Q40) for consensus.
Overall, CycloneSEQ data seems roughly comparable to ONT-fast data – CycloneSEQ was slightly better at consensus accuracy while ONT-fast was slightly better at read accuracy. The majority of CycloneSEQ’s consensus errors were homopolymer-length errors, often occurring in relatively short homopolymers (e.g. the ground-truth was G×5 but the assembly had G×4). This reminds me of ONT data from their previous pore (R9.4.1).
Basecalling greatly influences long-read accuracy, but CycloneSEQ’s basecalling process is unclear to me. Does CycloneSEQ offer different-sized basecalling models, similar to ONT’s fast/hac/sup? Are new models regularly released to allow for re-basecalling of existing data? Can users perform basecalling on a separate computer via the command line, or is it restricted to the workstation connected to the CycloneSEQ? I searched online for answers to these questions but found none. I only found mentions of CycloneMaster, a software tool that doesn’t appear to be freely available.
In conclusion, CycloneSEQ’s accuracy is not yet competitive with ONT. However, ONT’s accuracy improved dramatically over the course of its history, so I anticipate that CycloneSEQ may follow a similar trajectory. I will continue to watch this space!
Footnotes
-
Perhaps too much resemblance – Oxford Nanopore has filed a lawsuit. ↩
-
Transfer rates were extremely slow, and downloads frequently crashed. In order to get the three read files (two for short reads and one for long reads), I used
curlto download the files in 100 kB pieces (small enough to have a reasonable chance of being successful) and then stitched them together afterward. It took a couple of days and was not fun! Maybe CNGBdb were having technical problems that week? Or perhaps the performance is better from within China? ↩ -
Using the Achtman MLST scheme, our E. coli is ST1193 and the CycloneSeq E. coli is ST117. ↩
-
The CycloneSEQ data didn’t have any <1 kbp reads (presumably some length-based QC had already been applied) so my filter did nothing for that read set. ↩
-
Very low-quality reads are more likely to fail demultiplexing (i.e. go into the unclassified bin), so barcode-demultiplexed data tends to be a bit better on average than whole-flowcell data. ↩
-
Autocycler is the successor to Trycycler. At the time of writing, it’s not yet publicly released but will be very soon! ↩
-
I used
--carefulwith Pypolca. See our paper for lots of details on this polishing method: How low can you go? Short-read polishing of Oxford Nanopore bacterial genome assemblies. ↩ -
I used this script for comparing two alternative versions of an assembly. It gives the total difference count and shows the regions of difference in a human-readable manner. ↩
-
To get the mode, I rounded values to three significant figures and took the most common value. A subtle point: since qscore is a non-linear transformation of accuracy (-10 × log10(error rate)), the mean/mode of qscore-based accuracies is not equal to the mean/mode of percentage-based accuracies. So for this table, I calculated mean and mode separately for percentage-based and qscore-based accuracies. For anyone checking my maths, this is why mean/mode percentages and qscores seem to not match up – for example 90.9% equals Q10.4 not Q10.7. ↩
