A tale of two misassemblies
I’ve been getting a lot of mileage out of the nine ATCC genomes featured in some recent blog posts1. Currently, George Bouras and I are using them in a paper which examines low-depth Illumina polishing – stay tuned for that! While working on this paper, however, we saw two interesting misassemblies that I describe here. Both cases occurred with ONT-only assemblies (sup@v4.3.0 basecalling) performed by Flye.
Misassembly 1: Vibrio cholerae
The first misassembly happened with Vibrio cholerae ATCC-14035 (reads here). This genome has two circular chromosomes, the larger of which has an unusually long repeat (by bacterial standards) at 34 kbp.2 There are two exact copies of this repeat on opposite strands with 43 kbp of sequence in between. This configuration means that without any long reads to entirely span the repeat, the orientation of that middle 43 kbp would be unclear – it could point in either direction. Here is an illustration of the two configurations (not to scale):
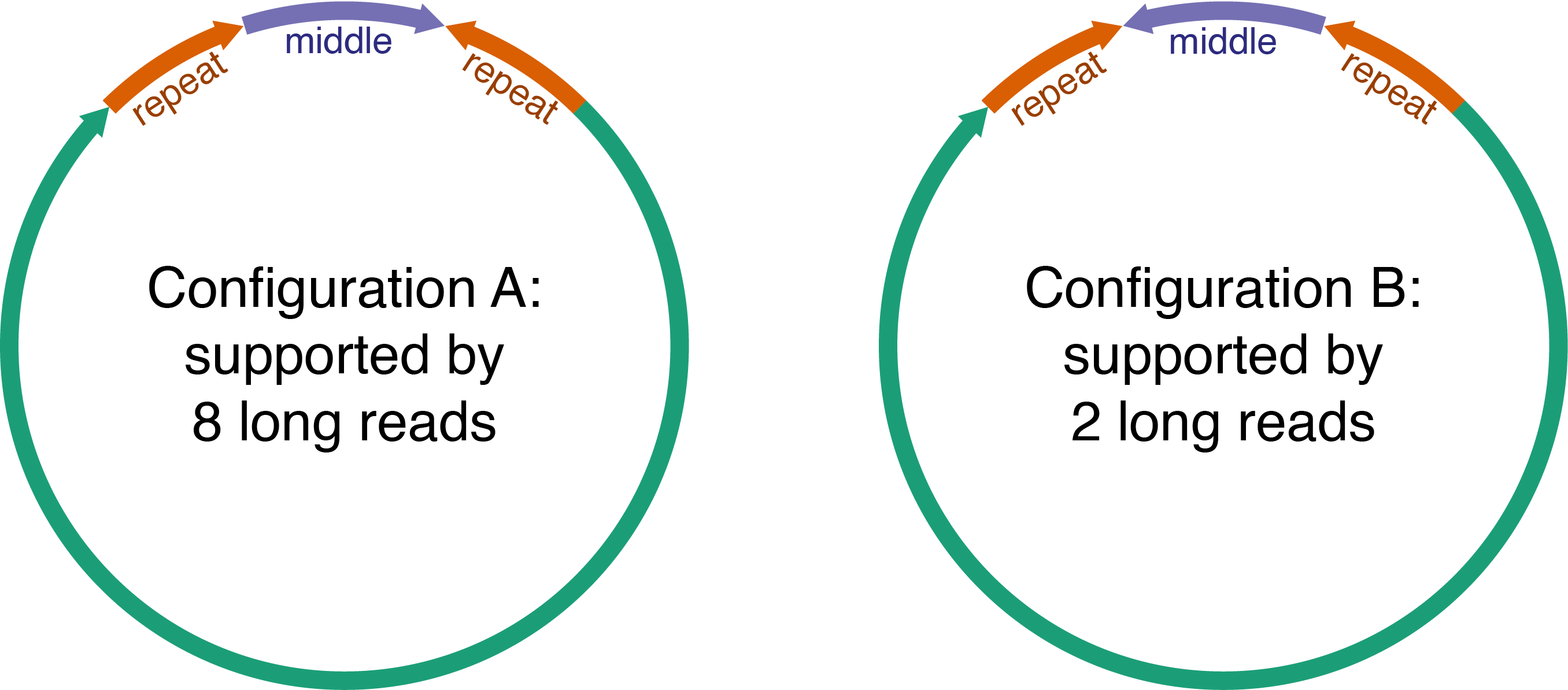
Thankfully, 10 reads do span the entire repeat, but they aren’t consistent: eight support configuration A and two support configuration B.3 It seems that a mixture of both configurations was in the sample. Perhaps homologous recombination occurred in the repeat, flipping the middle sequence around.
In cases like this, I would like an assembler to recognise the presence of heterogeneity and produce a contig consistent with the majority of the reads (configuration A).4 However, heterogeneity can confuse assemblers, and Flye chose to put both configurations in its contig, like this:

This is a linear contig with configuration A at the end and configuration B at the start. I can understand why Flye did this – it’s a single contig compatible with all of the reads. But it’s not an ideal outcome, as this contig includes extra sequence5 and has failed to circularise.
Misassembly 2: Listeria welshimeri
The second misassembly happened with Listeria welshimeri ATCC-35897 (reads here), where Flye deleted a 3956 bp chunk of the genome. George first noticed it when running hybracter (which uses Flye), and he saw that the deletion occurred about 70% of the time. I tried to replicate the problem on my computer, and I found it to depend on thread count: using 1 thread or 5+ threads resulted in a correct contig (no deletion) while using 2–4 threads resulted in the deletion.6
I have less to say about this misassembly because I cannot figure out why it happened. The deleted piece of the genome is not in a repetitive region (where misassemblies often occur), and I can’t spot any heterogeneity – all of the reads from this locus support the correct sequence and none contain the deletion. Very mysterious! It might require a deep dive into Flye’s algorithm to figure out what’s going on here.
Conclusions
This post is not meant to be a criticism of Flye. I quite like Flye – it’s one of my favourite long-read assemblers!7 But all assemblers, Flye included, make mistakes sometimes. So I wanted to remind readers that misassemblies can and do occur. Assemblies, especially those made with a single tool, should be viewed with some scepticism.
Structural heterogeneity is often the cause of misassemblies, because assemblers get confused with mixtures of different genome structures. If you were expecting a circular contig8 (i.e. the genome is circular) but got a linear contig, that is a clue that a misassembly due to structural heterogeneity may have occurred. But sometimes misassemblies have no obvious cause – they are simply a mistake made by the assembler.
Another interesting lesson is that assemblers can be non-deterministic: given the same input data, different runs may produce slightly different contigs.9 For some assemblers (including Flye), thread count can be a factor. In many scenarios, this non-determinism isn’t a problem – just don’t assume that re-running an assembly will produce the exact same result.
Finally, I’ll use this opportunity to once again plug my tool Trycycler. It is the best way I know of to avoid misassemblies like the two described in this post. Trycycler takes as input multiple separate assemblies of the same genome (ideally produced from different subsets of reads), and it combines them into a consensus assembly. Misassemblies can be caught at the reconciliation step (where they often create problems with circularisation) or at the consensus step (where only the majority variant at each locus is used). The caveat is that Trycycler usually takes some manual work, so it’s a slow way assemble a genome.
Footnotes
-
I first used these genomes here to look at accuracy after the move to 5 kHz sampling, and then again here after new Dorado basecalling models were released. ↩
-
George confirmed using pharokka that the repeat is prophage. ↩
-
Actually, these two reads are a duplex pair, i.e. they came from the two strands of a single piece of DNA. So there was only one sequenced DNA fragment supporting configuration B. ↩
-
Even better would be to include the most common configuration in the assembly but also add an annotation to describe the heterogeneity, but I’m not aware of any assembler that can do this. ↩
-
The correct counts for this genome are 2× repeat and 1× middle. The misassembled linear contig contains ~2.5× repeat and ~1.5× middle. ↩
-
The problem isn’t always due to thread count – George consistently used 4 threads and he sometimes got the deletion, sometimes did not. ↩
-
My other favourite is probably Canu, especially since I wrote this script to trim off start-end overlap in Canu’s circular contigs. Both Flye and Canu usually produce reliable assemblies, but Flye is faster so I use it the most. ↩
-
Different assemblers have different ways of indicating whether a contig is circular or linear. Some, like Flye, produce a GFA assembly graph that you can view in Bandage to see circularisation. Canu includes
suggestCircular=yesorsuggestCircular=noin contig header lines. Miniasm contig names end inc(for circular) orl(for linear). ↩ -
For short-read assemblies, SKESA is a tool which addresses this problem. It was specifically designed to be deterministic, regardless of thread count and read order. ↩
